Research lines
Below there is a brief description of the main research lines we are dealing with.
MS lesion segmentation in transversal studies
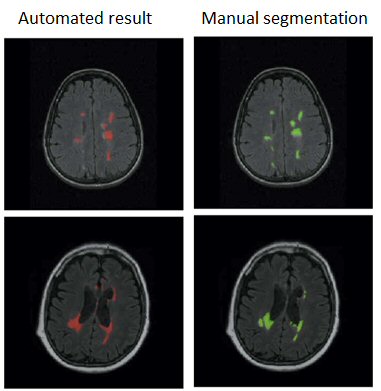
After a thorough analysis of the state-of-the-art on this topic, pointing out the importance of prior knowledge, and a subsequent review of atlas based segmentation of brain imaging, we have proposed different multiple sclerosis lesion segmentation pipelines.
The first one provides an initial tissue classification using a modified expectation-maximisation algorithm, which is later on refined with a lesion segmentation step based on thresholding and a region wise false positive reduction approach. The second one focuses only on the segmentation of lesions and uses an ensemble classifier alongside a rich feature pool including image intensities, probabilistic atlas maps, an outlier map and contextual information.
Longitudinal MS lesion detection and segmentation
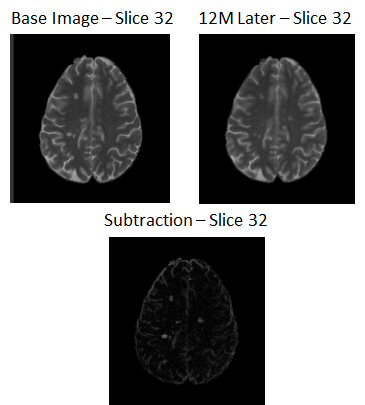
Time-series analysis of magnetic resonance images (MRI) is of great value for multiple sclerosis (MS) diagnosis and follow-up. We have presented an unsupervised subtraction methodology which incorporates multisequence information to deal with the detection of new MS lesions in longitudinal studies. The proposed pipeline consisted of the following steps: skull stripping, bias field correction, histogram matching, registration, white matter masking, image subtraction, automated thresholding, and postprocessing. We also combined the results of PD-w and T2-w images to reduce false positive detections.
The main challenges that voxel-to-voxel subtraction methods are dealing with are due to repositioning errors (patient movement), inconsistent objects over time such as blood and cerebrospinal fluid flow artifacts, noise in the images, and partial volume effects. Therefore, image registration, bias field correction, and intensity normalization are necessary steps to compensate for these problems.
Quantification of brain atrophy
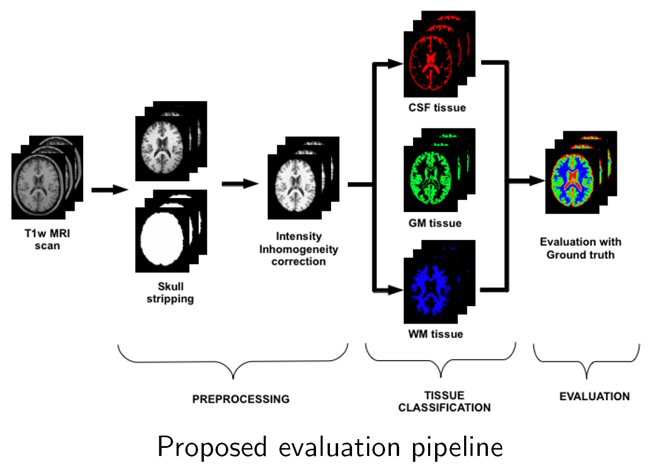
Brain atrophy can be quantified by comparing the difference of tissues in longitudinal analyses. However, tissue segmentation is influenced by white matter lesions (WML) if included in the segmentation process.
We have performed a quantitative evaluation of the impact of WML on different brain tissue segmentation methods. After evaluating the impact of WML on tissue volume estimation, we concluded that total grey matter volume is overestimated when lesion volume increased. The analysis done allowed us to prove that lesions have an important role on tissue volumetric measurement and led us to the development of a new pipeline for tissue segmentation in MS patients reducing the effect of the lesions. We developed first a new lesion-filling approach to improve the total tissue volume estimations and recently analysed in a breakthrough study the performance of fully automated tissue segmentation with both automatic lesion segmentation and lesion-filling methods.
Longitudinal non-rigid registration
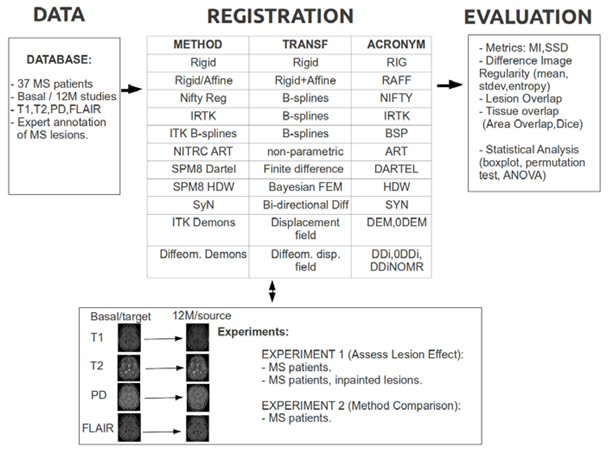
Registration is a key step in many automatic brain MRI applications. In this research line we focus on longitudinal registration of brain MRI for Multiple Sclerosis MS patients.
Our investigations show how a widely used method for longitudinal registration such as rigid registration is practically unconcerned by the presence of MS lesions while several non-rigid registration methods produce outputs that are significantly different.
In the Registration Toolbox we provide clues and links for the installation and testing of different deformable registration approaches. Part of the research done in this study has been applied to improve the detection of lesion changes in longitudinal studies.
Multimodal registration
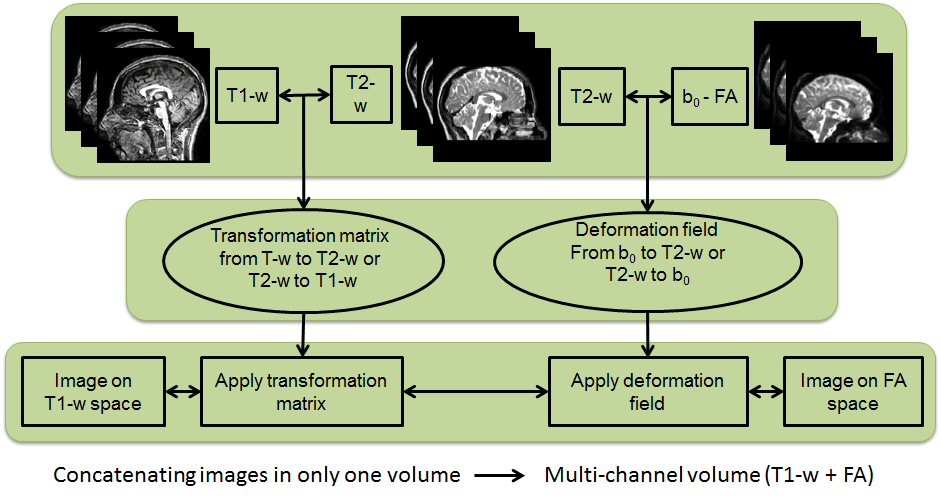
In this research line we analyse multi-channel registration approaches for different scans from people with MS. Such scans are affected by white matter lesions and there may also be marked brain tissue atrophy.
A particularly challenging task is to register T1w scans and diffusion weighted scans to a target space because of the geometrical distortions that affect the DW images and calculated maps. Multi-channel registration algorithms have the advantage of maintaining anatomical correspondence between T1w and DW images after registration to any target space. This is true even when large-scale deformations are necessary to match patients data to healthy targets.
